This product has been elaborated by Adam Master,
distributed by BioTe21 and presented as follows:
Mitochondrial DNA (mtDNA) is replicated inside of the mitochondrion as circular molecule of length of 16569 base pairs (rCRS, NCBI: J01415.2) and is inherited solely from the mother. A single mitochondrion contains 2 to 10 mtDNA copies, a single somatic cell (containing only two chromosome copies) has 100 - 10 000 mtDNAs, that illustrates its quantitative majority to genomic DNA present in a single copy per cell. This advantage is particularly significant for applications when using highly degraded biological material and was utilized in several studies into the mitochondrial DNA genotyping of Neanderthal fossils. The concept of the Mitochondrial Eve is also based on the mtDNA analysis, attempting to discover the origin of humanity by tracking the lineage back in time. This theory states that the mitochondrial DNA in all humans is inherited from one common female ancestor in Africa 150 000 - 200 000 years ago. Trace DNA analysis has become an integral part of a forensic laboratory's work and a key tool for investigators in forensic medicine. The low effective population size and rapid mutation rate (2-4% per million years) makes mtDNA also useful for assessing genetic relationships of individuals or population subgroups and also for identifying and quantifying the phylogeny (evolutionary relationships).The high mutation rate makes mtDNA most useful for comparisons of individuals within species.
As the individuals become more distantly related, the number of sequence differences becomes very large and changes begin to accumulate on changes until an accurate count becomes impossible. We are pleased to present complete systems and reagents allowing for their fast, cheap and reliable personal identification. Here, we would like to offer you mtDNAtest®-human - a Kit based on our optimized technology intended for human mitochondrial DNA analysis - hypervariable region I (HVI) and II (HVII). The Kit was designed to cover the major issues and challenges related to routine and reproductible trace DNA analysis in forensic sciences but is also useful in population genetics, tracing along a person's maternal line as well as personal identification on the basis of any biological materials.
PRINCIPLE
mtDNAtest®-human Kit allows for personal identification of humans and human remains, even when using higly degraded biological materials. Small nucleotide polymorphisms (SNPs) on the mitochondrial D-loop hypervariable regions can be also traced along a person's maternal line. The Kit is designed to help scientists in forensic medicine (comparative analysis of evidence samples and reference materials) medical diagnostics (identification of some cancer-related mutations in HVI and HVII regions of mtDNA) as well as other sciences (eg. migration analysis, pedigree generation for analysis of genetic linkage and association). The mtDNAtest®-human is based on higly sensitive PCR method intended for routine testing applications. Sequence analysis may be performed automatically with our special software: SNPtester® that allows you for identyfication of any SNP without knowledge about correct determination of sequence variations and usage of IUPAC nomenclature. Just put your row sequence in the SNPtester® and wait for the results.
Advantages of the Kit
- Simple and quick implementation of the procedures,
- Simple method, independent of laboratory practice,
- High yields even from higly-degraded samples
containing small-amounts of nucleic acids, - Standardized method for a variety of sample types,
- Optimized protocols for a range of starting materials,
- High sensitivity (DNA < 0.025 pg / reaction),
- Optimized PCR primer set,
- Excellent quality of results
(obtained amplicons do not cause problems during analyses), - Continuous help during analyses,
- Relatively low cost testing when compared to other methods.
Why mitochondrial instead of nuclear DNA?
- High copy number - quantitative majority - a single somatic cell contains ~ 10 - 1000 mitochondria and each mitochondrion carries 2 - 10 mtDNA copies, that result in about 100 - 10 000 copies of mitochondrial DNA per signle cell (containing nuclear DNA in one copy). This advantage is particularly significant for applications when using highly degraded biological material and was utilized in several studies into the mitochondrial DNA genotyping of Neanderthal fossils.
- High stability due to circular nature of mitochondrial DNA, making it easy to obtain for analysis, even from Neanderthal fossils.
- High mutation rate (10 times higher mutation rate than nuclear DNA), espcecially in hypervariable regions (HVI, HVII) acumulating small nucleotide polymorphisms (SNP) in a relatively small fragment of mitochondrial genome, that allows for comparisons of individuals and simplify analyses.
- Fixed in time mutation rate of mtDNA (2-4% per million years) - the fairly high mutation rate of mtDNA is reasonably fixed, making it useful to trace DNA changes through time of human evolution.
- No change in mtDNA from parent to offspring (lack of recombination) - maternal and paternal mtDNA usually do not recombine. Unlike nuclear DNA, which is inherited from mother and father and in which genes are frequently rearranged in the process of recombination, there is usually no change in mitochondrial DNA (mtDNA) from mother to offspring, keeping the mtDNA sequence clean. This makes mtDNA a powerful tool for genetic identification (particularly when genomic DNA is degraded).
- Maternal inheritance - human mtDNA is known for its maternal inheritance, that enables scientists to trace lineages back in time to the most recent common matrilineal (female) ancestor, often referred to as “mitochondrial Eve”, who was expected to live in Africa 150 000 - 200 000 years ago. The mtDNA studies place a population of a few thousand individuals in current Tanzania and close by regions at the base of the lineage that gave rise to modern man.
Why mtDNAtest®-human Kit ?
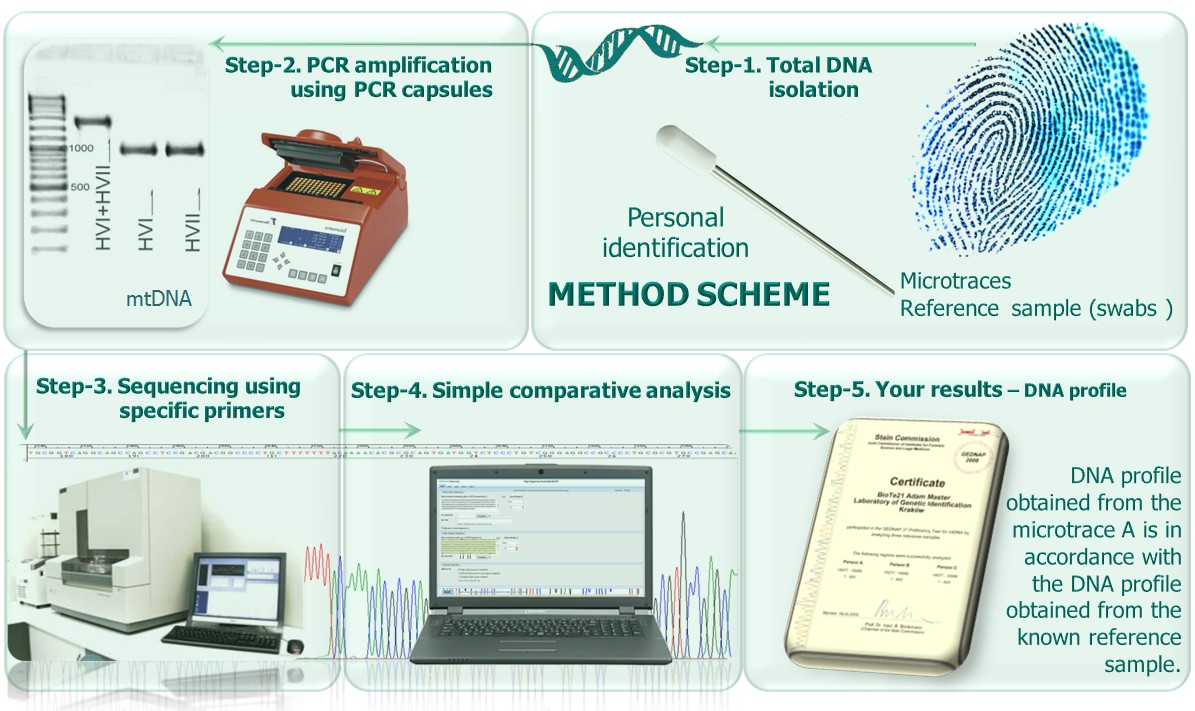
Both, many years of professional experience in the field of forensic molecular medicine, as well as the work with mtDNA allowed us to design and introduce a new test, which is, in technical terms, an extremely easy way to differentiate SNPs (Single Nucleotide Polymorphism) in mtDNA, based on a simple high-resolution sequencing method (seq module) or by using a capillary electrophoresis separation of fluorescently-labeled DNA fragments (shown above SNP-module, available soon). mtDNAtest®-human Kit allows you to use a variety of biological materials containing low DNA amounts (see sensitivity tests below). Selected and optimized PCR primer set allows for amplification of mtDNA D-loop sequence (see mtDNA scheme) comprising of whole hypervariable HVI and HVII sequences, therefore it is useful especially in genetic identification and comparative mtDNA analysis of biological traces.The The Kit is closed in delivered 0.2mL PCR tubes (as ready to use capsules), that limits potential contaminations of the reagents, ensures high repeatability of the results in routine work with trace DNAs and allows for shortening the whole identification process even up to 24h.
mtDNAtest®-human Kit sensitivity
High sensitivity of mtDNAtest®-human Kit (Primer Set B, for technical specification see below) enables fragment amplification of mtDNA hypervariable regions, even using very degraded trace samples. Only 0.00025ng of pure total DNA is needed to be added to one PCR reaction tube (referred to as "capsule") to obtain results as shown below:
Serial Dilutions of total human control mtDNA: 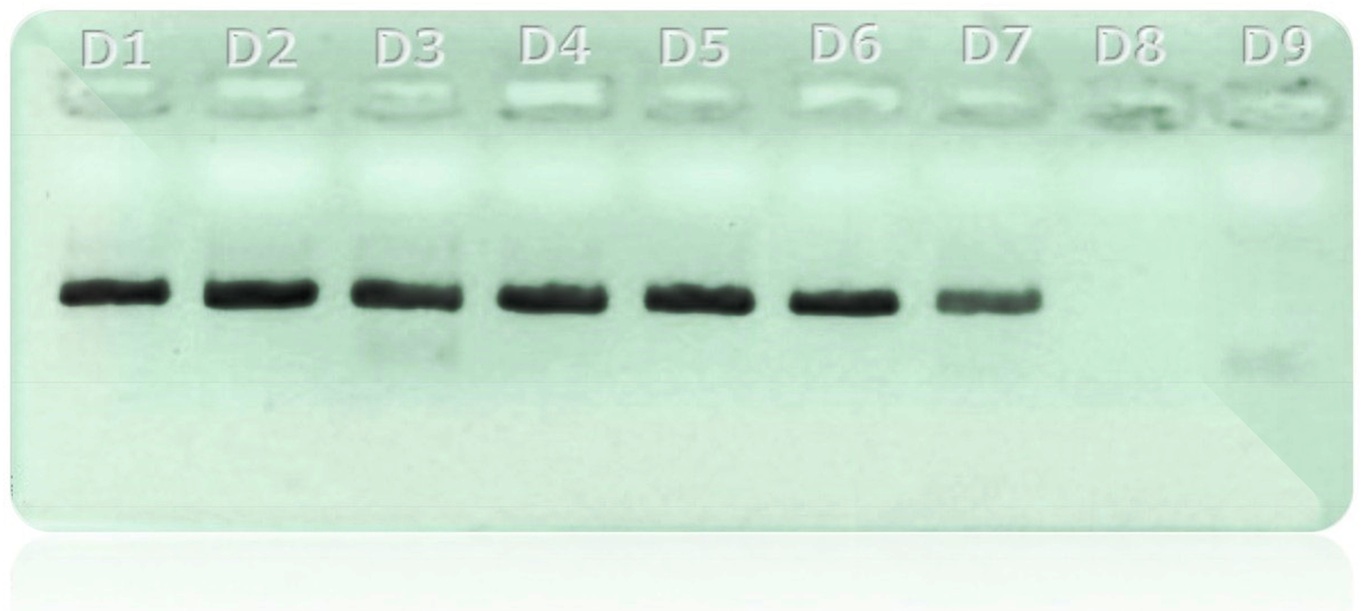
D1 25ng/PCR reaction
D2 2.5ng/PCR reaction
D3 0.25ng/PCR reaction
D4 0.025ng/PCR reaction
D5 0.0025ng/PCR reaction
D6 0.00025ng/PCR reaction
D7 0.000025ng/PCR reaction
D8 0.0000025ng/PCR reaction
D9 Negative control (H2O)
Using a range of control systems and prevent the formation of false negatives and false positives, you can provide clarity and certainty of your studies.
Analysis of mtDNA using this technology does not require any special laboratory adaptations and can be performed in any genetic laboratory dealing with direct PCR product sequencing and/or STR systems (called: Short Tandem Repeats) of genomic DNA, equipped with access to any thermocycler. The seq-module of the mtDNAtest®-human Kit operates using a variety of basic sequencing methods.
Validation: The technology proved their reliability during the international mtDNA quality tests (GEDNAP mtDNA – German DNA Profiling Group).
Materials & reagents
General information: mtDNAtest®-human seq-module is based on an optimized and validated PCR reagents that produce amplicons ready to use in direct sequencing of PCR products and further comparison with revised Cambridge Reference Sequence (rCRS, NCBI: J01415.2). Using mtDNAtest®-human seq-module you can obtain sequences that will allow you to score the sequence differences of your sample to the rCRS sequence.
The Kit includes reagents for PCR reactions and specific oligonucleotides for sequencing reactions. Optmized PCR reagents are closed in ready to use standard 0,2mL PCR tubes (referred to as capsules), which require only your template (total DNA) and an enzyme. Each PCR capsule contains a reaction mixture consisting of: an optimized PCR buffer, standarized PCR Primer Set, dNTPs, molecular biology grade water and other supplements improving the PCR reaction. Oligonucleotides for sequencing are provided in separate tubes.
For evaluation of your results, control DNA (positive control) and nuclease free water (non-template, negative control) are also provided.
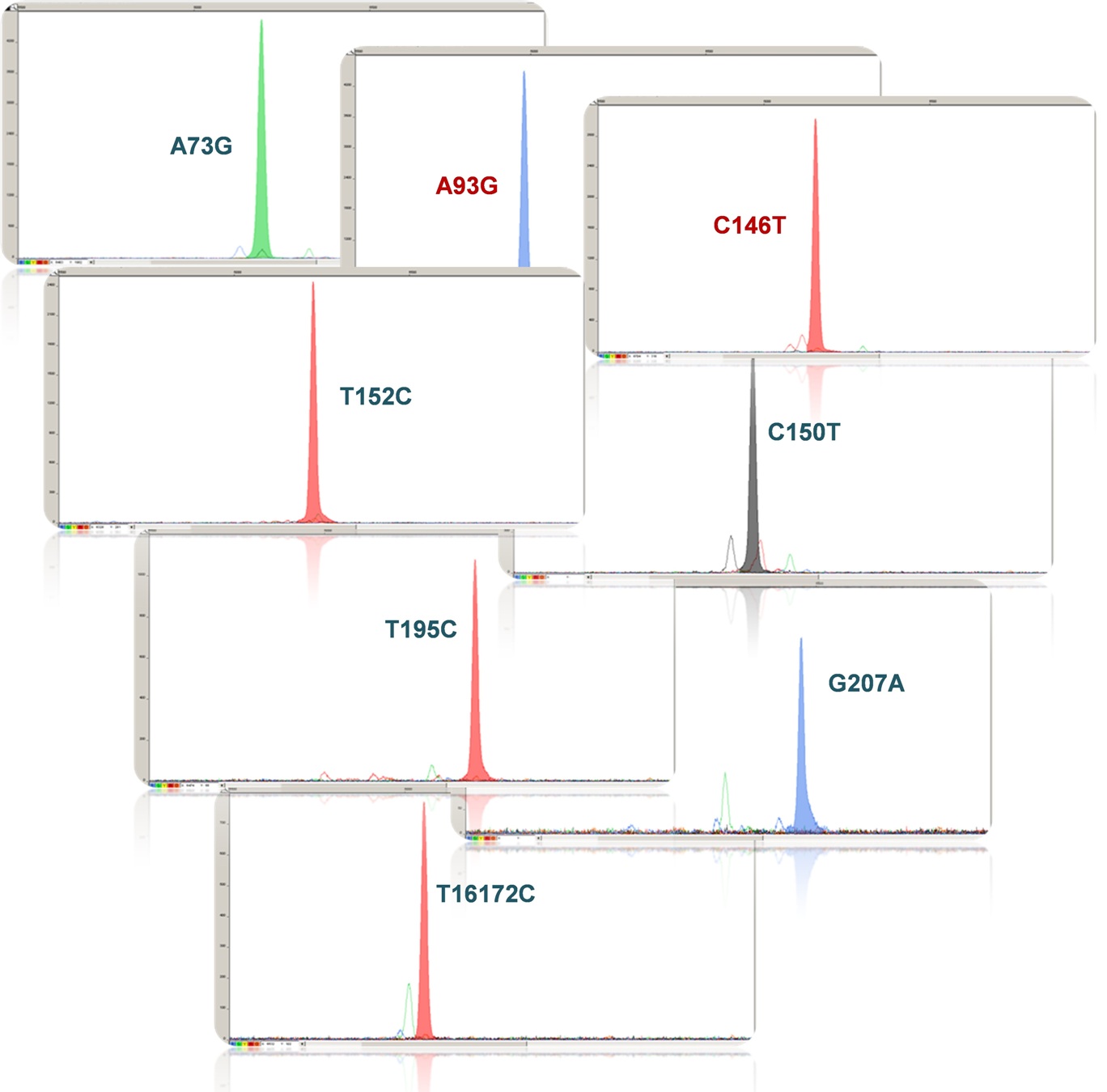
The control mtDNA (NCBI, GeneBank accession no.: KC342222), contains following SNPs:
HVI range: 16009-16569, SNP: T16126C, C16294T, C16296T, T16304C, A16463G, T16519C;
HVII range: 1-333, SNP: T3C, A73G, A263G, -309.1C, -315.1C
Equipment, reagents and materials required but not supplied:
You need only an enzyme 0,12uL/single capsule - a standard DNA polymerase with hot-start such as SuperHot Taq DNA Polymerase 5U/uL - Bioron (prefered) and typical laboratory equipment such as pipettes for the preparation of the PCR mixture and a thermocycler. If you have an instant access to any sequencer (genetic analyzer), you can perform all laboratory steps in 24h allowing for full identification of your samples in one day. There is also possibility to order the service (sequencing) in our lab or any other prefered lab. To sequence the PCR products in your laboratory you may need standard chemicals and materials used in the process (eg. BigDyeTerminator Sequencing Kit).
If you have any queries or problems with the technology, please do not hesitate to contact us.
PROCEDURE
mtDNAtest®-human seq-module
mtDNA testing is generally done in two regions of the genome known as hyper-variable regions: HVR1 (16024-16569) and HVR2 (00001-00576). Testing only HVR1 will produce low resolution results with a huge number of matches, so most experts generally recommend testing both HVR1 and HVR2 for more precise results. HVR1 and HVR2 test results also identify the ethnic and geographic origin of the maternal line.
If you've got deep pockets, a "full sequence" mtDNA test looks at the entire mitochondrial genome. Results are returned for all three regions of the mitochondrial DNA: HVR1, HVR2, and an area referred to as the coding region (00577-16023). A perfect match indicates a common ancestor in recent times, making it the only mtDNA test very practical for genealogical purposes. Because the full genome is tested, this is the last ancestral mtDNA test you will ever need to take. You may be waiting a while before you turn up any matches, however, because full genome sequencing is new enough that few people have yet to take the test.
It is important to understand that a full-sequence mtDNA test (but not the HVR1/HVR2 tests) may possibly provide information about inherited medical conditions - those that are passed down through maternal lines. If you don't want to learn this type of information, don't worry - it won't be obvious from your genealogy test report, and your results are well-protected and confidential. It would actually take some active research on your part or the expertise of a genetic counselor to turn up any possible medical conditions from your mtDNA sequence.
Polymerase Chain Reaction (PCR)
In the course of the analysis the mitochondrial DNA (mtDNA) replication is subjected to reaction using primers specific for the hypervariable region of mtDNA HVI. In the next stage, the resulting fragments are enzymatically cleaned. The resulting mixture of the matrix in the sequencing reaction. Identification using mtDNA amplification is based on the variable regions of the mitochondrial DNA (HVI), then sequenced using a specially selected set of oligonucleotides, in accordance with the given instructions. Separation of the products obtained by PCR is carried out during the electrophoresis in 1.5% agarose gel.
• PCR apparatus capable of carrying out the reaction in the PCR as described in section VII.1.; Genetic identification protocol using putty-animal mtDNAtest
• Sterile tubes for preparing mixtures for PCR.
11’ 95°C
- 25’’ 95°C
- 25’’ 59°C 45x
- 30’’ 72°C
15’ 73°C
4°C
Perform electrophoresis under the following conditions: 17 minutes with 180W.
We suggested that Samples prepared in this way can be sequenced with BigDye® Terminator v3.1 Cycle Sequencing Kit from Applied Biosystem on Applied Biosystems 3130/3130xl Genetic Analyzer.
From a technological point of view, for the proper PCR amplification of the analyzed material, important are the durability and integrity of the DNA. The amount of the output mtDNA copies has no greater influence on the STR analysis, than the amount of copies in gDNA (PCR reactions have got a fixed range of tolerance of the DNA quantity, regardless of its kind). However, a chance of an intact mtDNA copy preservation in a degraded material is much higher, which explains the superiority of the quantitative mtDNA over gDNA. Because of the above mentioned characteristics and its high durability, which is connected with the circular form of the cell, mtDNA is used in genetic identification and comparative analysis, especially when the analyzed biological material is highly degraded or when its amount is not sufficient to gDNA testing. The technology of mtDNA analysis is especially useful when analyzing: human corpses or their remains, microtraces like preserved clothes, cigarette stubs, and many more biological traces, for which the classic genomic DNA analysis is impossible. In such cases legal expert’s efficiency directly depends on the lab’s abilities in the field of alternative STR methods towards degraded gDNA material analysis.
Direct sequencing of PCR products
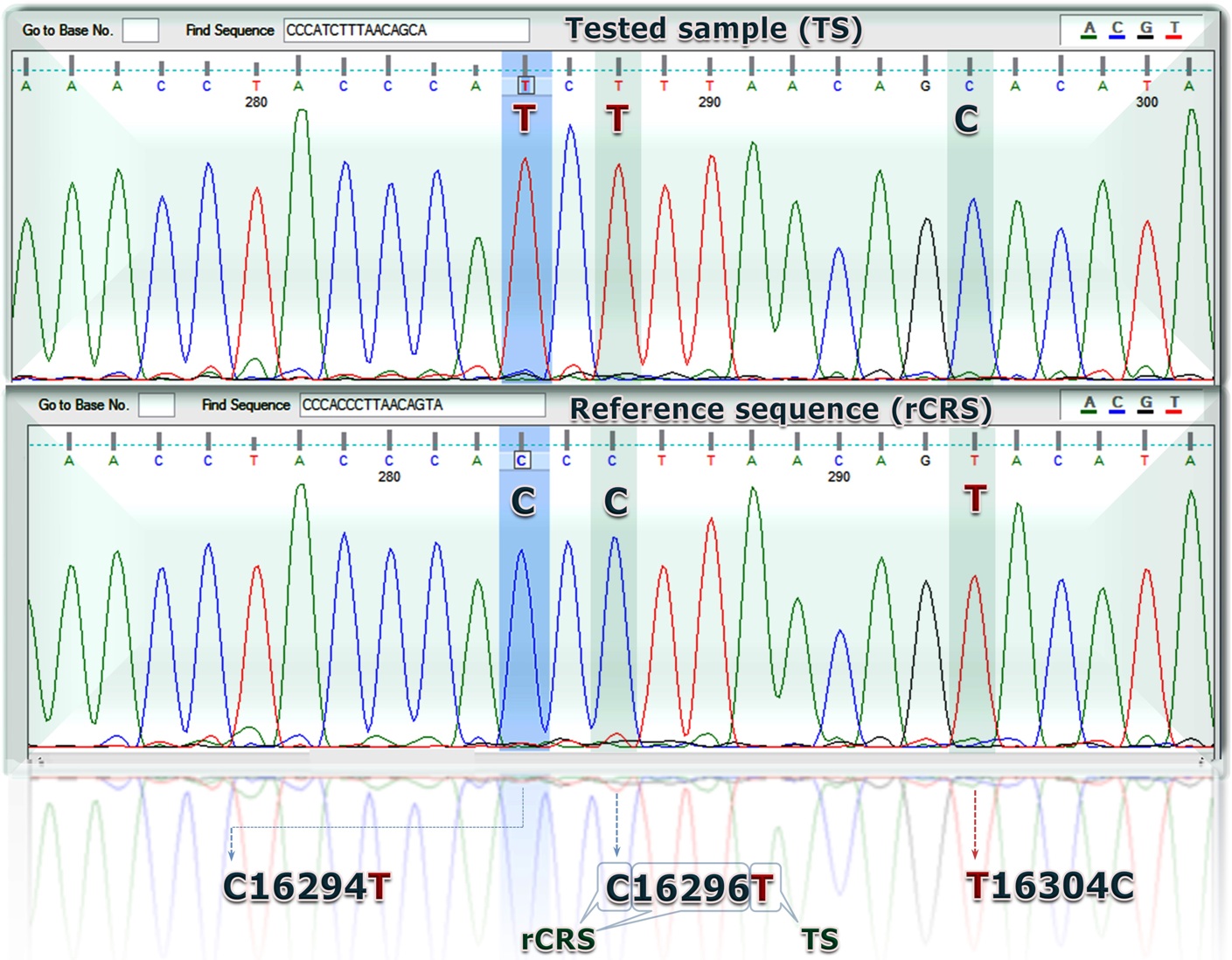
Your mtDNA results will generally be compared to a common reference sequence called the Cambridge Reference Sequence (CRS), to identify your specific haplotype, a set of closely linked alleles (variant forms of the same gene) that are inherited as a unit. People with the same haplotype share a common ancestor somewhere in the maternal line. This could be as recent as a few generations, or it could be dozens of generations back in the family tree. Your test results may also include your haplogroup, basically a group of related haplotypes, which offers a link to the ancient lineage to which you belong. Mutational events along the human mitochondrial DNA (mtDNA) phylogeny are traditionally identified relative to the revised Cambridge Reference Sequence (rCRS), a contemporary European sequence published in 1981. This historical choice is a continuous source of inconsistencies, misinterpretations and errors in medical, forensic and population genetic studies. Here, after having refined the human mtDNA phylogeny to an unprecedented level by adding information from 8,216 modern mitogenomes, we propose switching the reference to a Reconstructed Sapiens Reference Sequence (RSRS), which was identified by considering all available mitogenomes from Homo neanderthalensis.
Computational analysis
- Classical method - performed manually (see below) or
- With our special software: SNPtester® version 2.0, described on seperate page.
Final mtDNA profile usually consists of the sequence differences (mainly SNPs) in relation to the rCRS sequence. (eg.: ...C16296T, T16304C, A16463G,...) in which SNP numbers (...C16296T, T16304C, A16463G,....) represents a position on rCRS reference sequence, first letter position (...C16296T, T16304C, A16463G,...) referes to a nucleotide on rCRS reference sequence but second one (...C16296T, T16304C, A16463G,...)
first letter refered to

Results documentation
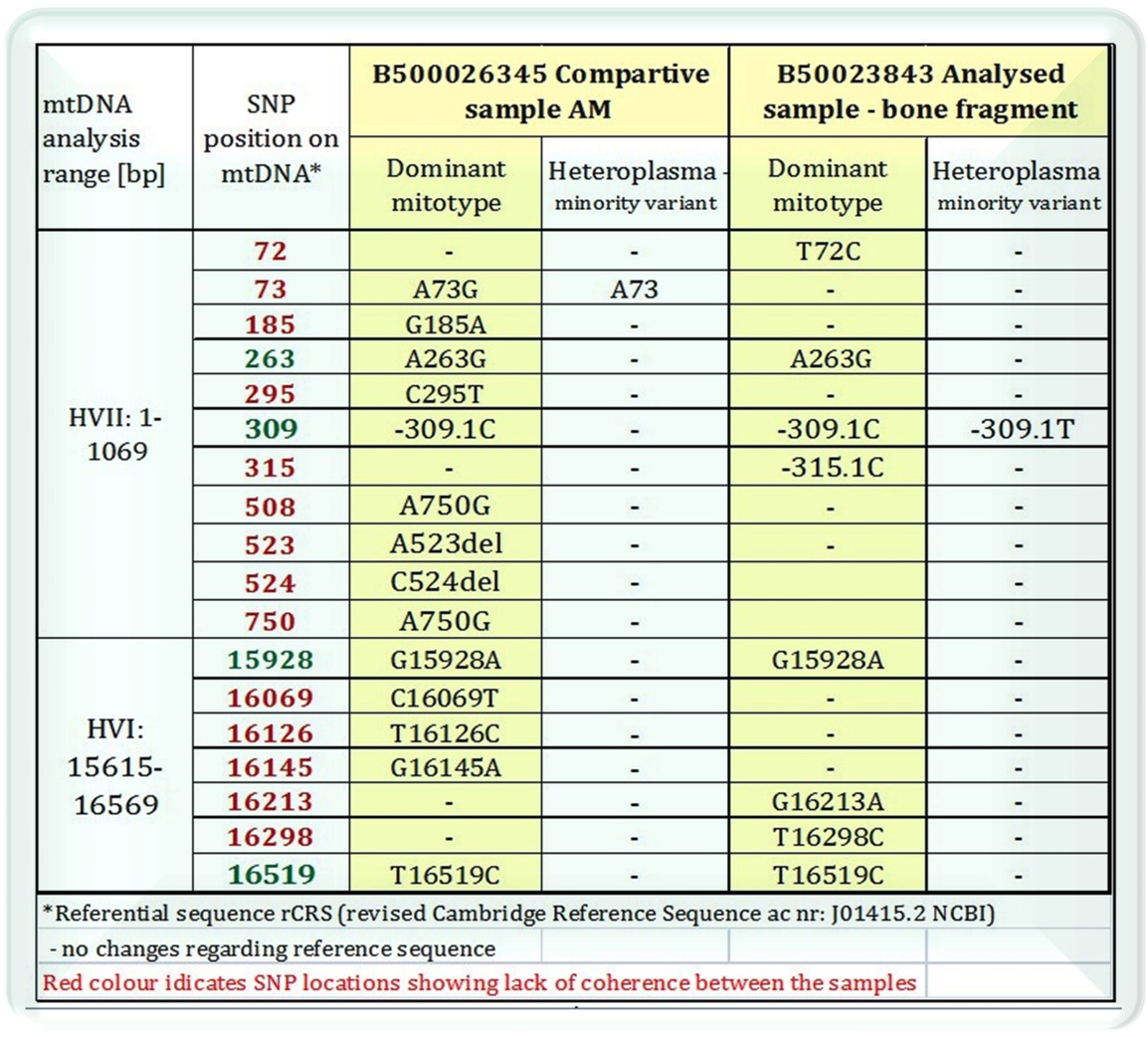
APPLICATIONS
mtDNAtest® -human module provide reliable results from laboratory analysis of forensic samples to personal identification. This product allows for fast and easy amplification of both hybervariable regions (whole D-loop sequence). The kit is also useful in population genetics, in the field of determining mtDNA haplogroups helpful in lineage analysis and family tree construction. Routine testing applications utilizing the Kit can be easily applied into any of your laboratory system as well as scaled up using automated workstations.
mtDNAtest® -human material and reagents are ready to use in all downstream assays, including applications in:
- Forensic science and medicine,
- PCR test detecting trace DNA (DNA isolates quality and quantity assesment)
- Human genetic testing research,
- Maternity testing,
- Cancer research
- Life science research,
- Routine applied testing,
- Pedigree genotyping.
Trace DNA analysis has become an integral part of a forensic laboratory's work and a key tool for investigators in forensic medicine. Accordingly, there has been considerable research conducted in order to provide the mtDNAtest®-human module as the best optimized technology to amplify trace mtDNA and improve its further analysis and interpretation. Although it is not possible to provide an exhaustive description of all applitactions and the numerous cases where trace DNA has provided key evidence, we hope that the Kit will gain the usefulness of trace DNA samples.
Introduction to mtDNA analysis
Mitochondrial DNA (mtDNA) is replicated inside of the mitochondrion as circular molecule of length of 16569 base pairs (Revised Cambridge Reference Sequence - rCRS, NCBI: J01415.2) and is inherited solely from the mother. mtDNA includes both 37 genes (13 protein coding, 22 tRNA genes) as well as non-coding regions, and highly variable HVI and HVII sequences (see figure below). Mitochondrial DNA is inherited throughout the maternal line, therefore its analysis allows, among others, finding mother-children natural cognation, as well as relationship between supposed siblings such as brother-sister, grandmother-grandchildren in maternal line (paternally inherited Y-chromosomal DNA is used in an analogous way to trace the agnate lineage). Some of the identification may be performed only using mtDNA analysis, useful, for example, in identification of deceased mother of siblings, as well as performing a comparative identification analysis of even highly degraded corpses and bone fragments. mtDNA analysis is also performed instead of STR genotyping method, when genomic DNA (gDNA) is highly-degraded.
A single mitochondrion contains from 2 to 10 mtDNA copies, a single somatic cell (containing only two chromosome copies) contains from 100 to 10 000 mtDNAs, an egg contains 100 000 to 1 000 000 mtDNA molecules, whereas a sperm contains only 100 to 1000, that illustrates its quantitative majority to genomic DNA present in a single copy per cell. This may also explain why sperm is inherited only from the mother. The low effective population size and rapid mutation rate makes mtDNA useful for assessing genetic relationships of individuals or population subgroups and also for identifying and quantifying the phylogeny (evolutionary relationships). The concept of the Mitochondrial Eve is based on the mtDNA analysis, attempting to discover the origin of humanity by tracking the lineage back in time. To do this, scientiscs determine and then compare the mtDNA sequences from different individuals from all over the world. In humans and animals, the high mutation rate makes mtDNA most useful for comparisons of individuals within species. As the individuals become more distantly related, the number of sequence differences becomes very large; changes begin to accumulate on changes until an accurate count becomes impossible.
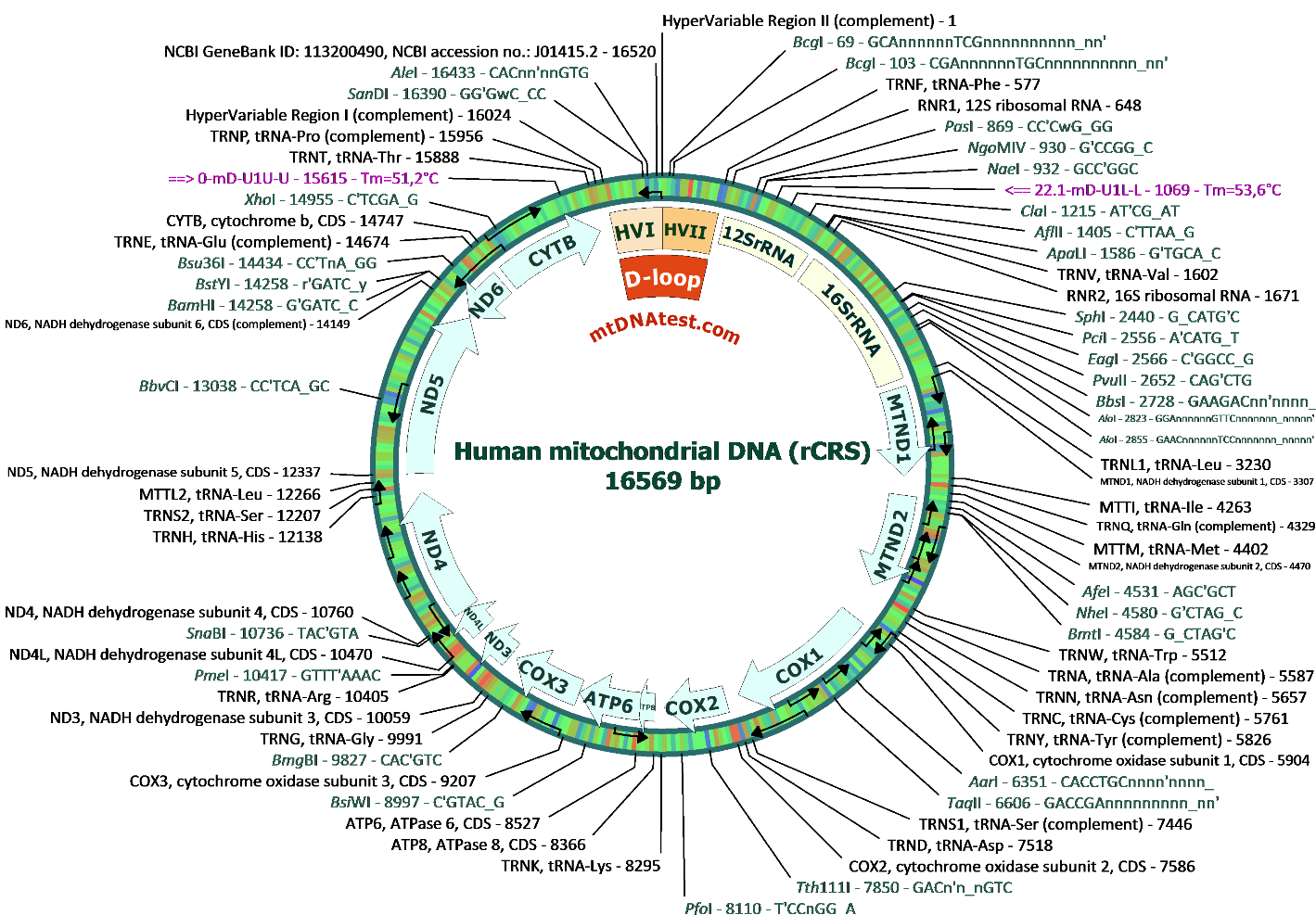
Mitochondrial DNA map of rCRS
Fig.1. Map of human mitochondrial genome prepared on the basis of mtDNA reference sequence: db_xref GeneBank ID: 113200490, NCBI accession no.: J01415.2 . This Revised Cambridge Reference Sequence (rCRS) has eighteen specific corrections or confirmations of the original 1981 sequence of Anderson et al [PMID:7219534]. Positions of consensus sequences recognized by restriction endonucleases (1 cutters) were shown by dark green font. mtDNA features (including rRNA, tRNA and protein coding regions were idicated by black font. The most important regions for mtDNA function were also shown inside the circle as gene names or other recognizable names such as HVI, HVII (hypervariable region I and II) in D-loop sequence flanked by distal mtDNAtest primer set marked in purple (pDRAW 32). The various circle colours (blue-green-red) were shown to indicate different melting temperatures of separate mtDNA regions. More mtDNA features were presented on the high resolution mtDNA figure.
References
-
Marquez MC. Interpretation guidelines of mtDNA control region sequence electropherograms in forensic genetics. Methods Mol Biol. 2012;830:301-19. doi: 10.1007/978-1-61779-461-2_21. PubMed PMID: 22139669.
-
Bandelt HJ, van Oven M, Salas A. Haplogrouping mitochondrial DNA sequences in Legal Medicine/Forensic Genetics. Int J Legal Med. 2012 Nov;126(6):901-16. doi: 10.1007/s00414-012-0762-y. Epub 2012 Sep 1. PubMed PMID: 22940763.
-
Seo Y, Uchiyama D, Kuroki K, Kishida T. STR and mitochondrial DNA SNP typing of a bone marrow transplant recipient after death in a fire. Leg Med (Tokyo). 2012 Nov;14(6):331-5. doi: 10.1016/j.legalmed.2012.06.001. Epub 2012 Jul 7. PubMed PMID: 22776743.
-
Salas A, Coble M, Desmyter S, Grzybowski T, Gusmão L, Hohoff C, Holland MM, Irwin JA, Kupiec T, Lee HY, Ludes B, Lutz-Bonengel S, Melton T, Parsons TJ, Pfeiffer H, Prieto L, Tagliabracci A, Parson W. A cautionary note on switching mitochondrial DNA reference sequences in forensic genetics. Forensic Sci Int Genet. 2012 Dec;6(6):e182-4. doi: 10.1016/j.fsigen.2012.06.015. Epub 2012 Jul 25. PubMed PMID: 22840856.
-
Holland MM. Molecular analysis of the human mitochondrial DNA control region for forensic identity testing. Curr Protoc Hum Genet. 2012 Jul;Chapter 14:Unit14.7. doi: 10.1002/0471142905.hg1407s74. PubMed PMID: 22786611.
-
Rizzi E, Lari M, Gigli E, De Bellis G, Caramelli D. Ancient DNA studies: new perspectives on old samples. Genet Sel Evol. 2012 Jul 6;44:21. doi: 10.1186/1297-9686-44-21. Review. PubMed PMID: 22697611; PubMed Central PMCID: PMC3390907.
-
Green RE, Briggs AW, Krause J, Prüfer K, Burbano HA, Siebauer M, Lachmann M, Pääbo S. The Neandertal genome and ancient DNA authenticity. EMBO J. 2009 Sep 2;28(17):2494-502. doi: 10.1038/emboj.2009.222. Epub 2009 Aug 6. Review. PubMed PMID: 19661919; PubMed Central PMCID: PMC2725275.
-
Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc Biol Sci. 2009 Apr 7;276(1660):1201-9. doi: 10.1098/rspb.2008.1758. Epub 2009 Jan 20. Review. PubMed PMID: 19203921; PubMed Central PMCID: PMC2660971.
- Chinnery PF. Inheritance of mitochondrial disorders. Mitochondrion. 2002 Nov;2(1-2):149-55. PubMed PMID: 16120317.
-
Saneto RP, Sedensky MM. Mitochondrial Disease in Childhood: mtDNA Encoded. Neurotherapeutics. 2012 Dec 6. PubMed PMID: 23224691.
-
Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013 Jan 15;22(2):384-90. doi: 10.1093/hmg/dds435. Epub 2012 Oct 16. PubMed PMID: 23077218; PubMed Central PMCID: PMC3526165.
-
Pakendorf B, Stoneking M. Mitochondrial DNA and human evolution. Annu Rev Genomics Hum Genet. 2005;6:165-83. Review. PubMed PMID: 16124858.
-
Disotell TR. Discovering human history from stomach bacteria. Genome Biol. 2003;4(5):213. Epub 2003 Apr 28. Review. PubMed PMID: 12734001; PubMed Central PMCID: PMC156578.
Useful links
Personal identification
- Haplogrouping mitochondrial DNA sequences in Legal Medicine/Forensic Genetics
- Interpretation Guidelines of mtDNA Control Region Sequence Electropherograms in Forensic Genetics
- The causes of mutation accumulation in mitochondrial genomes
Genetic genealogy
- The DNA Ancestry Project
- Begin your ancestral journey
- DNA reveals Neanderthal extinction clues - BBC
- Ancient DNA studies: new perspectives on old samples
- Mitochondrial DNA Clarifies Human Evolution
- How Mitochondrial Eve connected all humanity and rewrote human evolution
- Mitochondrial Eve - Mother of All Humans Lived 200,000 Years Ago
- Ancient DNA and Neanderthals
- The Neandertal genome and ancient DNA authenticity
- Neanderthals Didn't Mate With Modern Humans, Study Says
- Taking the mtDNA pulse of Neandertal populations
- Discovering human history from stomach bacteria
Inheritance of mitochondrial disorders
- Mitochondrial inheritance
- Mitochondrial Disorders Overview by NCBI
- mtDNA and Mitochondrial Diseases
- Mitochondria research society
- Mitochondrial Disease in Childhood: mtDNA Encoded
- Mitochondrial Mutations Make Tumors Spread
- Universal heteroplasmy of human mitochondrial DNA
As other IVD, the Kit is intended for applications utilizing molecular biology methods. This product is neither intended for the diagnosis, prevention, or treatment of a disease, nor has it been validated for such use either alone or in combination with other our products.