Advances in understanding the molecular basis
of cancer pathogenesis promise to improve the detection, diagnosis, and treatment of these diseases.
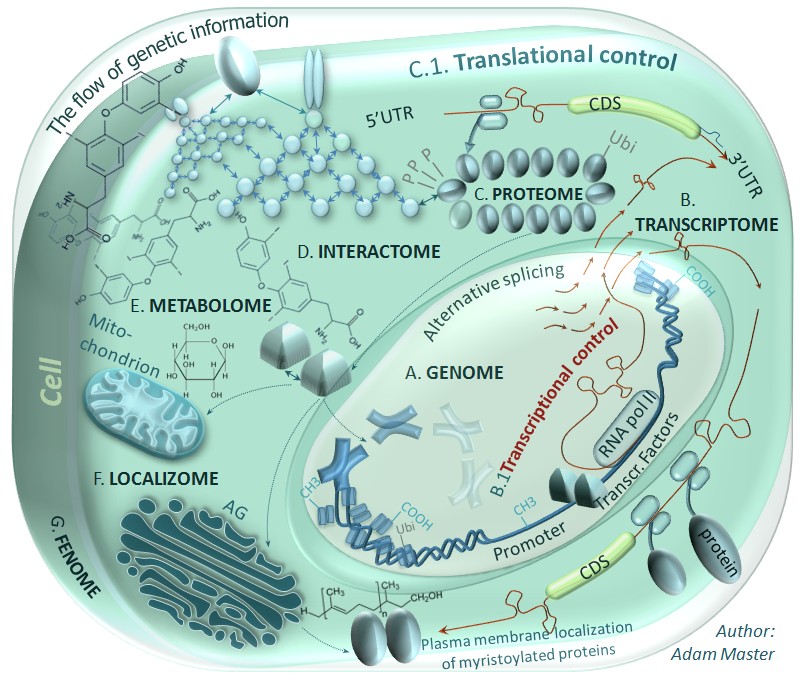
The sequencing of whole genomes revealed that most of the human DNA is transcribed, and less than 2% of the human genome codes for proteins, which means that the majority of the transcripts are non coding RNAs (ncRNAs), and their function remains to be established. Moreover, the new technologies of the whole genome sequencing and screening of cDNA libraries showed that about 20% of human and mouse genes overlap resulting in formation of potential sense-antisense pairs. Though, the physiological relevance of most of the pairs remains obscure, there are a number of identified and annotated antisense transcripts that are known to regulate expression of genes involved in cell growth, proliferation, differentiation, apoptosis, metabolism and circadian cycle.In contrast to a group of short non-coding RNAs (sncRNAs) such as microRNA, the long naturally occuring antisense transcripts (NATs) including long non-coding RNAs (lncRNAs) usually exhibit the higher complementarity, and hence the higher specificity towards target genes.
 The NATs are able to form sense-antisense pairs showing the potential to activate mechanisms controlling target gene expression. Thus these naturally occuring moleculaes are emerging as important targets for new therapeutics.
The NATs are able to form sense-antisense pairs showing the potential to activate mechanisms controlling target gene expression. Thus these naturally occuring moleculaes are emerging as important targets for new therapeutics.
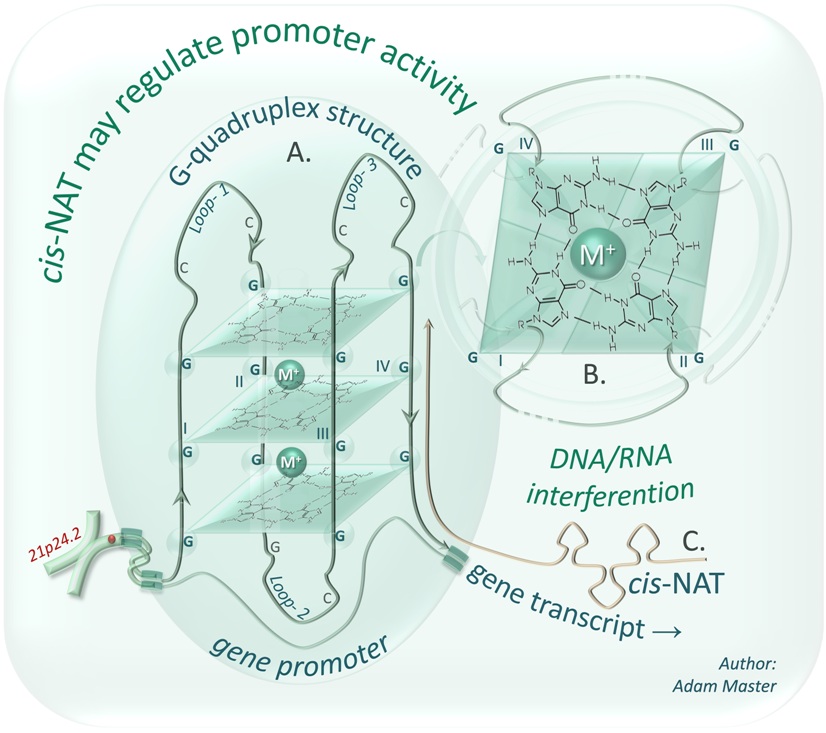
My current work on this topic focuses on research aimed at understanding the role of selected long naturally occuring antisense transcripts (NATs) in carcinogenesis as well as on elaboration of alternative strategies for controling the NATs and their target genes' expression in cancer cells. Surprisingly, recent studies have shown that interfering RNAs may also activate gene transcription via newly discovered phenomenon of small RNA-induced gene activation (RNAa). The small activating RNAs (saRNAs) have been demonstrated as promoter-specific activators enhancing transcription efficiency of target genes. However, our results showed that at least some published saRNAs could be in fact siRNAs silencing NATs that are complementary to sense strand of their target genes' promoters. The scheme (below) may be an example of one of such regulations, in which a NAT transcript interferes with a sense DNA strand forming an inhibitory G-quadruplex structure. This interference may change the target gene transcription, however the DNA/RNA interaction may be blocked by the use of siRNA technology that results in degradation of the cis-NAT.
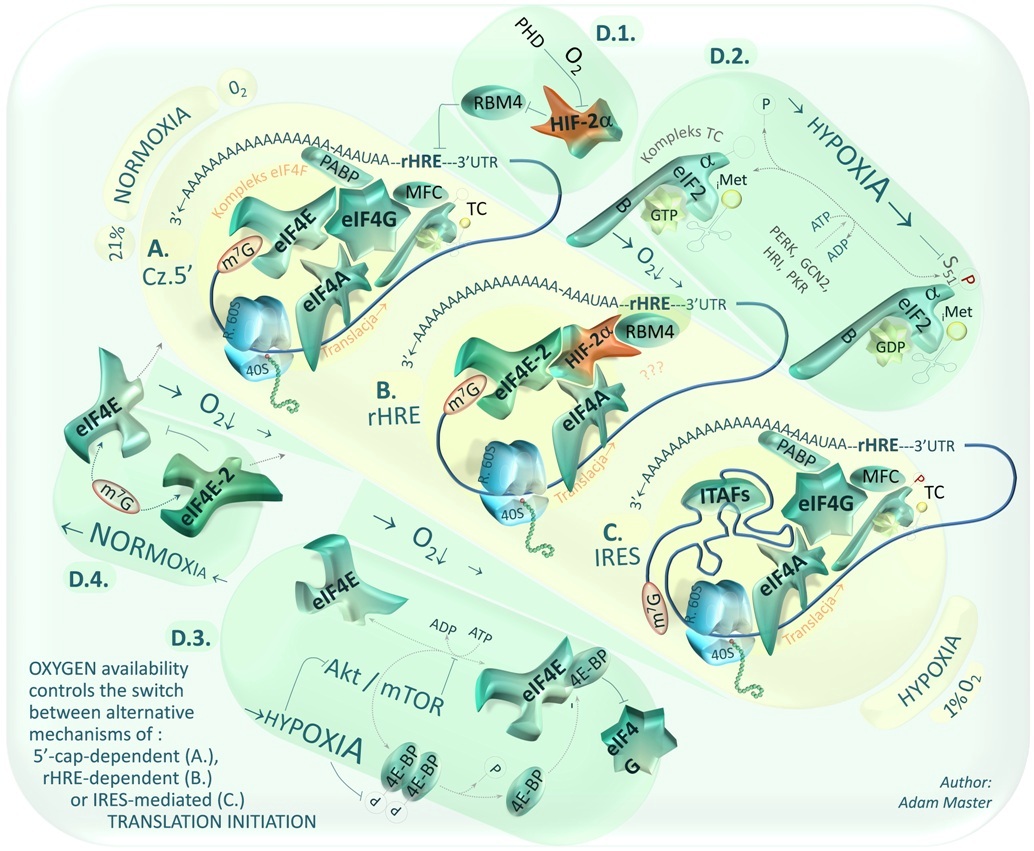
 Translational control has been found to be aberrant in the course of various diseases including cancers and neurodegenerative disorders, whose pathogeneses are poorly understood. This control is a mechanism of protein synthesis regulation emerging as an important target for new therapeutics. Thus there is the need for further research to determine the roles of cis- and trans-acting factors controlling alternative mechanisms of translation initiation in both physiology and pathology of various tissue types. Binding of these factors to DNA or RNA may obstruct regulatory elements or influence the stability of some functional structures such as RNA G-quadruplexes or internal ribosome entry sites (IRES elements) and finally silence (e.g. with microRNA, siRNA) or enhance (saRNA) oncogene's or suppressor's expression. At present, my research on translational control is focused on 5'UTR-mediated regulation of protein biosynthesis and in particular on enhancement of expression of selected suppressors. Schemes (above, on the right) presents some of eucariotic initiation factors' -mediated control of protein biosynthesis and alternative mechanisms of translation initiation that may be switched during hypoxia as a result of cellular integrated stress response (ISR) induced by low oxigen levels.
Translational control has been found to be aberrant in the course of various diseases including cancers and neurodegenerative disorders, whose pathogeneses are poorly understood. This control is a mechanism of protein synthesis regulation emerging as an important target for new therapeutics. Thus there is the need for further research to determine the roles of cis- and trans-acting factors controlling alternative mechanisms of translation initiation in both physiology and pathology of various tissue types. Binding of these factors to DNA or RNA may obstruct regulatory elements or influence the stability of some functional structures such as RNA G-quadruplexes or internal ribosome entry sites (IRES elements) and finally silence (e.g. with microRNA, siRNA) or enhance (saRNA) oncogene's or suppressor's expression. At present, my research on translational control is focused on 5'UTR-mediated regulation of protein biosynthesis and in particular on enhancement of expression of selected suppressors. Schemes (above, on the right) presents some of eucariotic initiation factors' -mediated control of protein biosynthesis and alternative mechanisms of translation initiation that may be switched during hypoxia as a result of cellular integrated stress response (ISR) induced by low oxigen levels.
 In my work with Human Thyroid Hormone Receptor Beta gene (THRB suppressor), and CDKN2A I have demonstrated for the first time that the newly discovered phenomenon of small RNA-induced gene activation (RNAa) may also enhance protein translation by acting at sequence-specific targets within the mRNA 5’-untranslated region (5’UTR). Short oligodeoxynucleotides (termed dGoligos) targeting of the TRß1 5'UTR markedly increased the efficiency of the TRß1 protein translation showing a therapeutic potential of sense/antisense based dGoligos as Gibb's free energy modulators of 5'UTR stem-loop structure. The dGoligos, as a set of oligonucleotide-based trans-acting factors was designed, specifically targeting alternatively spliced 5’UTRs (see figure above) in transcripts expressed from the TRß1. For instance, the in vitro translation efficiency of reporter constructs containing alternative TRß1 5’UTRs was increased by up to more than 6.5-fold following exposure to specific oligonucleotides. These results would be the first showing a method for gene-specific translation enhancement using selective trans-acting factors designed to target specific 5’UTR cis-acting elements of high regulatory potential (report under editorial consideration).
In my work with Human Thyroid Hormone Receptor Beta gene (THRB suppressor), and CDKN2A I have demonstrated for the first time that the newly discovered phenomenon of small RNA-induced gene activation (RNAa) may also enhance protein translation by acting at sequence-specific targets within the mRNA 5’-untranslated region (5’UTR). Short oligodeoxynucleotides (termed dGoligos) targeting of the TRß1 5'UTR markedly increased the efficiency of the TRß1 protein translation showing a therapeutic potential of sense/antisense based dGoligos as Gibb's free energy modulators of 5'UTR stem-loop structure. The dGoligos, as a set of oligonucleotide-based trans-acting factors was designed, specifically targeting alternatively spliced 5’UTRs (see figure above) in transcripts expressed from the TRß1. For instance, the in vitro translation efficiency of reporter constructs containing alternative TRß1 5’UTRs was increased by up to more than 6.5-fold following exposure to specific oligonucleotides. These results would be the first showing a method for gene-specific translation enhancement using selective trans-acting factors designed to target specific 5’UTR cis-acting elements of high regulatory potential (report under editorial consideration).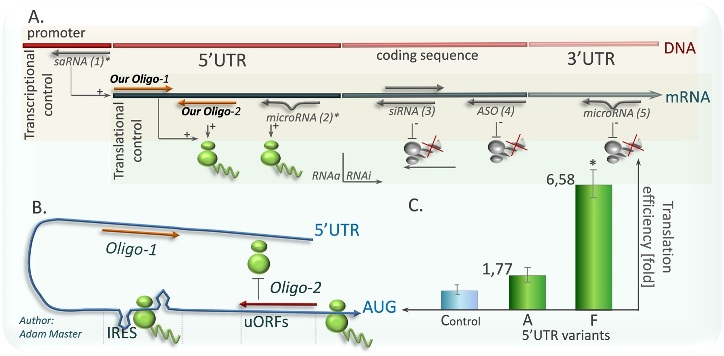
Preliminary results of the translation enhancement of the TRß1 was raported on 34th Annual Meeting of the European Thyroid Association (Master et al 2009, ETA, OP18) and 17th Annual Meeting of the American Society of Gene & Cell Therapy (Master et al. 2014, poster). This simple strategy may be developed in the future to complement other available methods for gene expression regulation including gene silencing. The oligonucleotide-mediated enhancement of protein expression has the potential to be transferred to increase the translation efficiency of any suitable target gene and may have future application in gene therapy strategies to enhance expression of proteins such as tumor suppressors.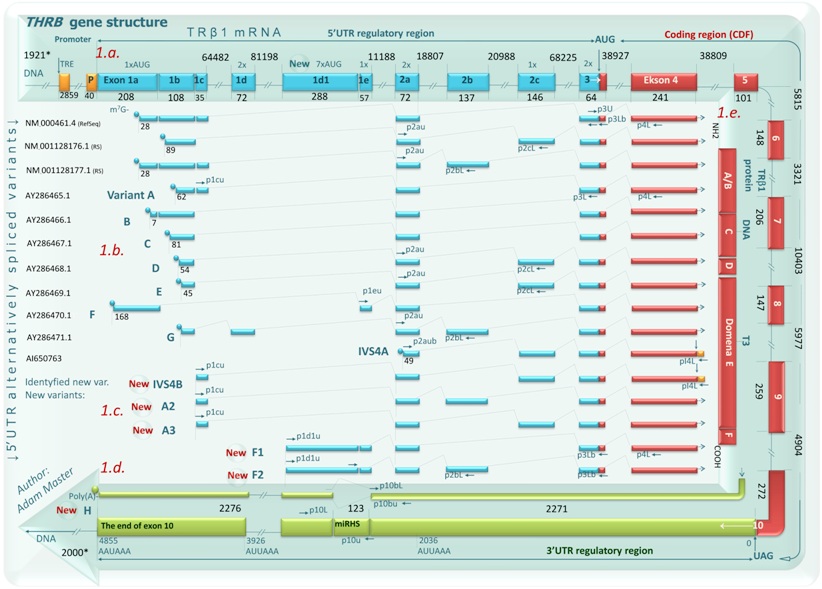
In my PhD studies on the role of mRNA UTR regions in translational control of thyroid hormone receptor beta 1 (TRß1) in human clear cell Renal Cell Carcinoma (ccRCC), I have shown that the expression of alternatively spliced 5’-UTR mRNA variants of the TRß1 is dependent on the tissue type and aberrant in ccRCC. I have also identified 4 new (not reported before) 5’UTR variants of the TRß1 mRNA (F1, F2, A2, A3) expressed in normal kidney and ccRCC, a new variant encoding truncated protein (IVSB) and a new 3’UTR variant that lacks a discrete region containing putative binding sites for miRNAs (splice variant H, see figure). Two variants, F1 and F2, contained a previously unknown exon (1d1) located between exons 1d and 1e (Master at al 2010). Moreover, our study showed that the aberrant expression of TRß1 5'UTR variants together with altered levels of expression of TRß1 binding miRNAs in ccRCC have been demonstrated to be responsible for the poor correlation between mRNA and protein levels, thus providing compelling evidence for UTR-dependent regulation of TR protein synthesis in ccRCC (Master at al 2010).
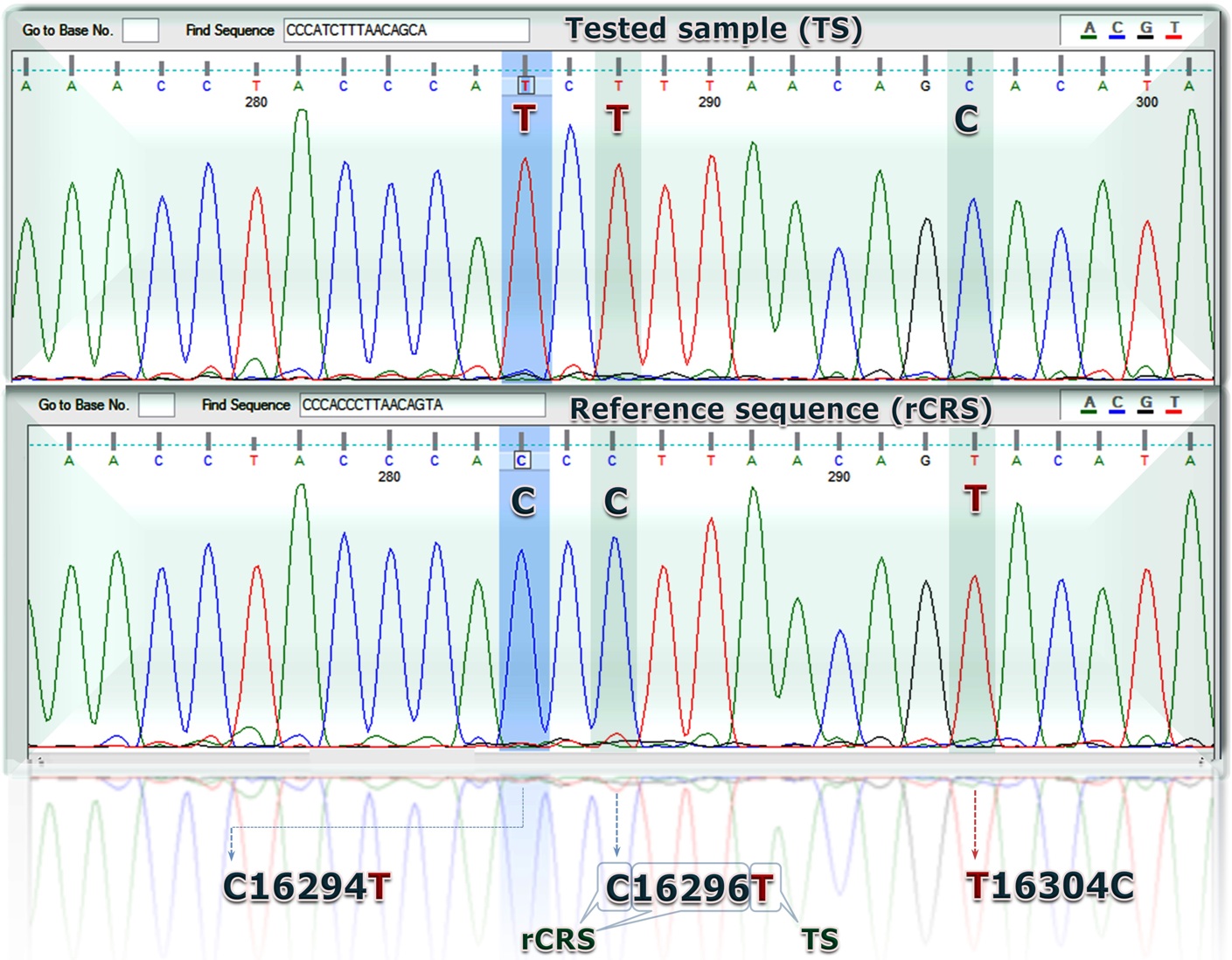 As it was shown in this research, ccRCC may be characterized by local tumor hypothyroidism resulting from low level of tissue T3, TRβ1 and DIO1 that may be involved in the carcinogenesis process itself or in maintaining a proliferative advantage to the malignant cells on thyroid hormone receptor β1 (TRβ1) acts as a ligand dependent transcription factor whose activity is regulated by thyroid hormone, triiodothyronine (T3). The study has also shown that translational control via UnTranslated Regions of mRNAs may have an important role in key physiological pathways for cell proliferation, metabolism and responses to cellular stresses that may have a direct impact on cancer development and progression (Master at al 2010).
As it was shown in this research, ccRCC may be characterized by local tumor hypothyroidism resulting from low level of tissue T3, TRβ1 and DIO1 that may be involved in the carcinogenesis process itself or in maintaining a proliferative advantage to the malignant cells on thyroid hormone receptor β1 (TRβ1) acts as a ligand dependent transcription factor whose activity is regulated by thyroid hormone, triiodothyronine (T3). The study has also shown that translational control via UnTranslated Regions of mRNAs may have an important role in key physiological pathways for cell proliferation, metabolism and responses to cellular stresses that may have a direct impact on cancer development and progression (Master at al 2010). 
New published sequences (NCBI GeneBank Acc.no.):
GQ456950 Homo sapiens thyroid hormone receptor beta 1 mRNA, 5' UTR, alternatively spliced;
5' UTR splice variant F1 - 5'UTR fragment containing: newly discovered exon 1d1, and 1e,2a and 3.
GQ456951 Homo sapiens thyroid hormone receptor beta 1 mRNA, 5' UTR, alternatively spliced
5' UTR splice variant F2 - 5'UTR fragment containing: newly discovered exon 1d1 and 1e,2a,2b and 3.
GQ456948 Homo sapiens thyroid hormone receptor beta 1 mRNA, 5' UTR, alternatively spliced;
5' UTR splice variant A2 - 5'UTR fragment containing: exon 1c,2a, 2b and 3.
GQ456949 Homo sapiens thyroid hormone receptor beta 1 mRNA, 5' UTR, alternatively spliced;
5' UTR splice variant A3 (5'UTR fragment containing: exon 1c,2a, 2c and 3.
GQ869479.1 Homo sapiens thyroid hormone receptor beta 1 mRNA,
5' UTR splice variant A4, alternatively spliced.
GQ919288.1 Homo sapiens truncated thyroid hormone receptor beta 1
5' UTR splice variant IVS4B (THRB) mRNA, complete cds, alternatively spliced.
GQ456952.1 Homo sapiens thyroid hormone receptor beta 1 mRNA, 3' UTR, alternatively spliced.
3'UTR splice variant H splice variant H.
GQ869477.1 Homo sapiens thyroid hormone receptor beta 1 mRNA,
3' UTR splice variant 1, alternatively spliced.
GQ869478.1 Homo sapiens thyroid hormone receptor beta 1 mRNA,
3' UTR splice variant 2, alternatively spliced.
KC342222.1 Homo sapiens isolate 20121113-Kr1 tRNA-Pro (TRNP) gene and D-loop,partial sequence; mitochondrial.
KC342221.1 Sus scrofa domesticus isolate 20121206-11 cytochrome b (CYTB) gene, partial cds; tRNA-Thr and tRNA-Pro genes,
complete sequence; and D-loop, partial sequence; mitochondrial
KC332217.1 Humulus lupulus voucher 20121204-847b 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S
ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 26S ribosomal RNA gene, partial sequence
KC292629.1 Cannabis sativa subsp. indica voucher 20121121-846 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S
ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 26S ribosomal RNA gene, partial sequence
and the other sequences: FJ002249.1, FJ002248.1, FJ002247.1, FJ002246.1, FJ002245.1, FJ002244.1, FJ002243.1 of Homo sapiens
iodothyronine deiodinase type 1 splice variant a mRNA, complete cds, alternatively spliced (second author).
List of published sequences.