Applying innovative technologies to the analysis
Applying innovative technologies to the analysis
of genetic variation and function.
RNAi > siRNA > RNAa > saRNA > ASO > miRNA > RNA/DNA synthesis >
new bio-farmaceutics based on nucleic acid molecule >
DNA/RNA analysis > gDNA > mtDNA > SNP > STR > sequencing > epigenetics > microarray > laboratory diagnostics >
Engineering recombinant-protein > genetic constructs > protein synthesis & purification > bio-farmaceutics >
Elaborated products:
Medical laboratory diagnostics allowing for fast and routine screening for:
-
Increased risk of developing UV-dependent melanoma malignum (MC1R, CDKN2A),
-
Hereditary Prostate, Breast and Ovarian Cancer (BRCA1, BRCA2, CHEK2, NBS1),
-
Factor V Leiden (SNP: R506Q; F5) prothrombin (SNP: G20210A; F2),
-
and many others (based on SNP/mutation detection).
Melanoma malignum
melTest Kit® is a unique product used in UV-dependent skin cancer (malignant melanoma) predisposition diagnosis. melTest® has been designed thanks to many years of experience in the field of genetic diagnostics. This allowed me to create a test specifically designed to detect predisposition for UV-dependent skin cancer. The diagnostics of UV-dependent skin cancer is based on DNA sequencing technology, as well as fluorescent analysis of SNP (mini-sequencing). An analysis of up to 15 SNP-type mutations in MC1R gene and up to 6 mutations in CDKN2A gene (both in correlation with high risk of skin cancer occurrence) is possible.
Using the technology, there is no need for special preparation or adaptation of the laboratory. Analysis may be performed in any genetic laboratory (equipped with a thermocycler and a sequenator) specialized in testing using SNP or STR (Short Tandem Repeats) analysis of genomic DNA.
Understanding the information about mutations associated with some hereditary diseases, and using preventive genetic diagnostics, may help the individuals in changing their life-styles (especially risky behavior, like an extensive sun-light exposition) if they have UV-dependent skin cancer predisposition, and therefore, save their lives.
Hereditary Breast and Ovarian Cancer

BRCA2: Lys3326X, Tyr3401Met,Tyr3401Met, G8410A (Val2728Ile), C5972T (Thr1915Met), 4088delA (ter1291), G7985A (Trp2586Ter), 488delCT, G4486T, A1093TC,
NBS1: C643T, R215W, 657del5.
CHEK2: 1100delC, I157T, IVS2+1G>A, del5395, Glu239Lys (G715A), Glu239Stop (G715T).


Selective enhancement of protein synthesis by small synthetic RNA termed eRNA
The technology complements genes silencing methods (siRNA, miRNA). However, what makes eRNA different and exceptional, is the fact, that instead of inhibiting, it enhances the translational efficiency of chosen genes in selective manner. Both in biology and molecular medicine, selective silencing of genes is already being performed by the means of: 1) synthetic siRNA or antisense molecules as well as 2) non-selective, native cellular miRNAs. However, until now, there was no method for enhancement of selected genes.
Just like siRNA (artificial small inhibitory RNA) and miRNA (naturally occurring inhibitory microRNA), the eRNA may use RNAi related phenomena and/or RNAse H pathways to induce cellular response. However, what makes the eRNA different and exceptional, is the fact, that instead of inhibiting it leads to protein synthesis enhancement of selected genes or gene fragments even by 500%, in selective manner.

eRNA in the context of other oligonucleotide-based trans-acting factors that can trigger mechanisms of RNA interference (RNAi) or RNA activation (RNAa).
Some properties of the eRNA:
• acts as a protein synthesis enhancer at the level of translational control,
• does not change in significant way the level of transcription,
• targets a selected mRNA - as designed before on the basis of a chosen sequence of a single gene or even its allele characterized by a single nucleotide polymorphism (SNP).

Some practical uses for eRNAs:
1. enhancement of suppressors, mutators, modulators and other genes involved in anti-cancer response: (TP53, BAX, CDKN2A, CDK4, RB1, NOD2, CHEK2, INBS1, XPD, MC1R, MSH2, MLH1, MSH6, BRCA1, BRCA2, APC and others),
2. enhancement of anti-viral response by activation of: interferons (IFNA1, IFNA2, IFNB, IFNG) and others,
3. enhancement of anti-aging genes such as mutator genes, telomerase (TERT), collagens: (COL1A1-COL9A1) and others that activation maybe useful in aesthetic medicine,
4. enhancement of genes involved in hereditary diseases,
5. enhancement of genes, that expression is too low or is wanted to be higher, including cytokines and growth factors (Il1-Il10, VEGF, TGF, TNF, EGF, FGF), trans-membrane receptors, protein hormones (INS, GH1, IGH), proteins of signaling pathways, apoptotic pathways, enzymes and other genes important for genetic studies and industrial biotechnology,
6. enhancement of selected splicing variants of a single gene (for example: the longer variant of the BRCA1 gene),
7. selective enhancing of a correct allele of a gene in heterozygous cells (SNP-differing gene alleles wherein one of the alleles is mutated, the other one is correct),
8. overexpression of a selected gene at the level of translation,
9. studies on cell cycle, proliferation, growth, differentiation, embryogenesis, carcinogenesis and apoptosis, endocytosis, exocytosis, internalization and intercellular communication,
10. studies on interaction between proteins,
11. enhancing translational efficiency in technological processes of recombined proteins,
12. and many more that we can you propose you on the basis of your studies.
Genetic identification
Description: The kit allows for personal identification of humans and human remains, even when using higly degraded biological materials. Mutations/SNPs on the mitochondrial DNA D-loop hypervariable regions can be traced along a person's maternal line.
Purposes: forensic (comparative analysis of evidence samples and reference materials) medical (identification of some cancer-related mutations in HVI and HVII regions of mtDNA), scientific (migration analysis).
Test type: higly sensitive PCR.
Description: The kit allows for genetic identification of any mammal species (cat, dog, horse, cow, pig, deer, fox etc.) and other animals, including many birds, amphibians and reptiles, as well as individuals within a species (individual animal identification). The samples are usually of unknown origin from: accidents involving animals, illegal hunting, bites, pedigree analysis, and identification of animal droppings in a public space.
Purposes: veterinary, forensic, scientific.
Test type: higly sensitive PCR.
Description: The kit allows for genetic identification of any plant and fungus genus and species in a sample containing cultured or uncultured bacteria.
Purposes: medical (swabs of pathogenic fungi), forensic (fungi comparison), scientific , industrial (foodstuffs).
Test type: fast, cheap, highly sensitive PCR.
Description: The kit allows for genetic identification of any bacteria genus or species in a sample containing cultured or uncultured bacteria.
Purposes: a) medical (pathogenic bacteria) especially difficult to culture anaerobic bacteria, b) forensic, c) scientific.
Test type: fast, cheap, higly sensitive PCR.
mtDNAtest is an optimized technology for mitochondrial DNA analysis and its implementation.The unique mtDNAtest kit was designed for the purpose of genetic identification and mtDNA genotype determination. The kit is useful in population genetics, in the field of determining haplotypes and haplogroups helpful in lineage analysis and family tree construction.
Years of professional experience in the field of forensic molecular medicine, as well as the work with mtDNA allowed me to design and introduce a new test, which is, in technical terms, an extremely easy way to differentiate SNPs (Single Nucleotide Polymorphism) in mtDNA, based on the method of fluorescent DNA fragments separation (mtDNAtest-SNP module). mtDNAtest kit will allow to yield reliable results, without the need of difficult sequence analysis. Specially chosen set of analyzed SNP allows analyzing the most hyper variable HVI and HVII sequences’ locations, therefore it is useful especially in genetic identification and comparative mtDNA analysis of biological traces.

Apart from the mentioned innovative test of specific SNPs, it can be used a standard kit for the variable mtDNA regions’ sequencing with specially chosen set of oligonucleotides, allowing to yield highest quality results every single time (mtDNAtest-SEK module).

mtDNA analysis, with the help of the technologies, does not require a specially equipped lab and, therefore, may be performed in any genetic laboratory routinely analyzing STR (Short Tandem Repeats) of genomic DNA (gDNA). All you need is a thermocycler and a standard sequenator. The above presented mtDNAtest kit’s modules use different basic methods (minisequencing or classic sequencing) and may be employed separately.
Elaborated software:
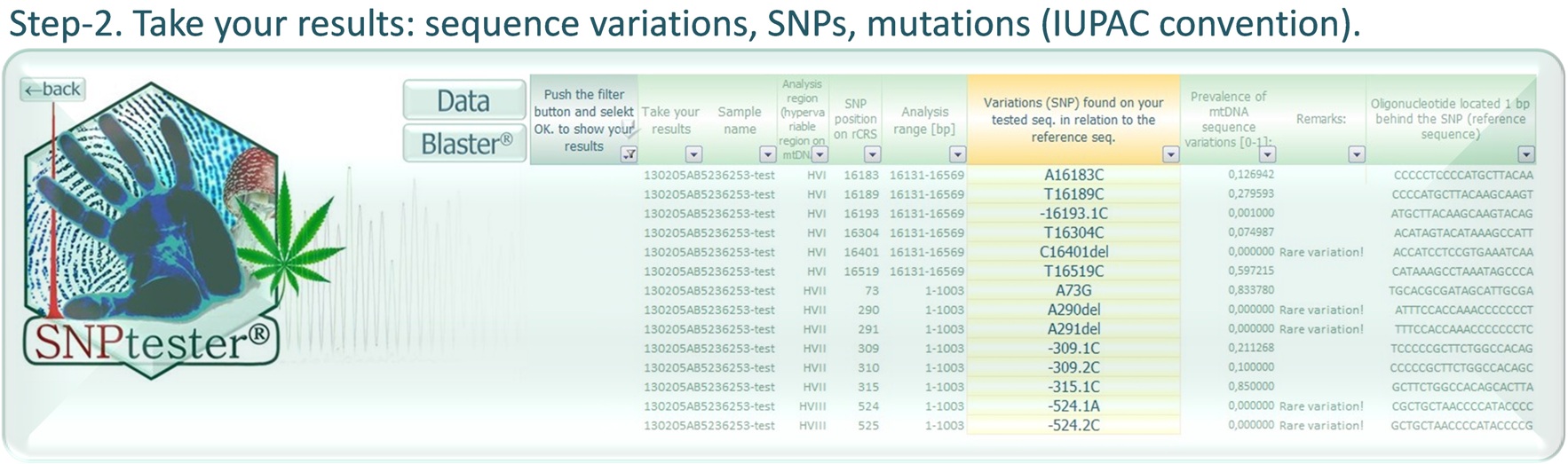
SNPtester® version 2.0 is a macro working with Microsoft Excel, that is intended for analysis of sequence variations. It allows you for quick and reliable identification of small nucleotide polymorphisms (SNP), termed according to the IUPAC convention. Originally, it was designed to simplify sequence analysis of mitochondrial DNA (mtDNA), however it works with any sequence that can be compared with any reference sequence. The SNPtester® is an extremely simple tool to help you improve your sequence analysis, allowing for fast identification of sequence variations as well as more consistent and correct usage of unequivocal nomenclature.
Sequence analysis in three steps:
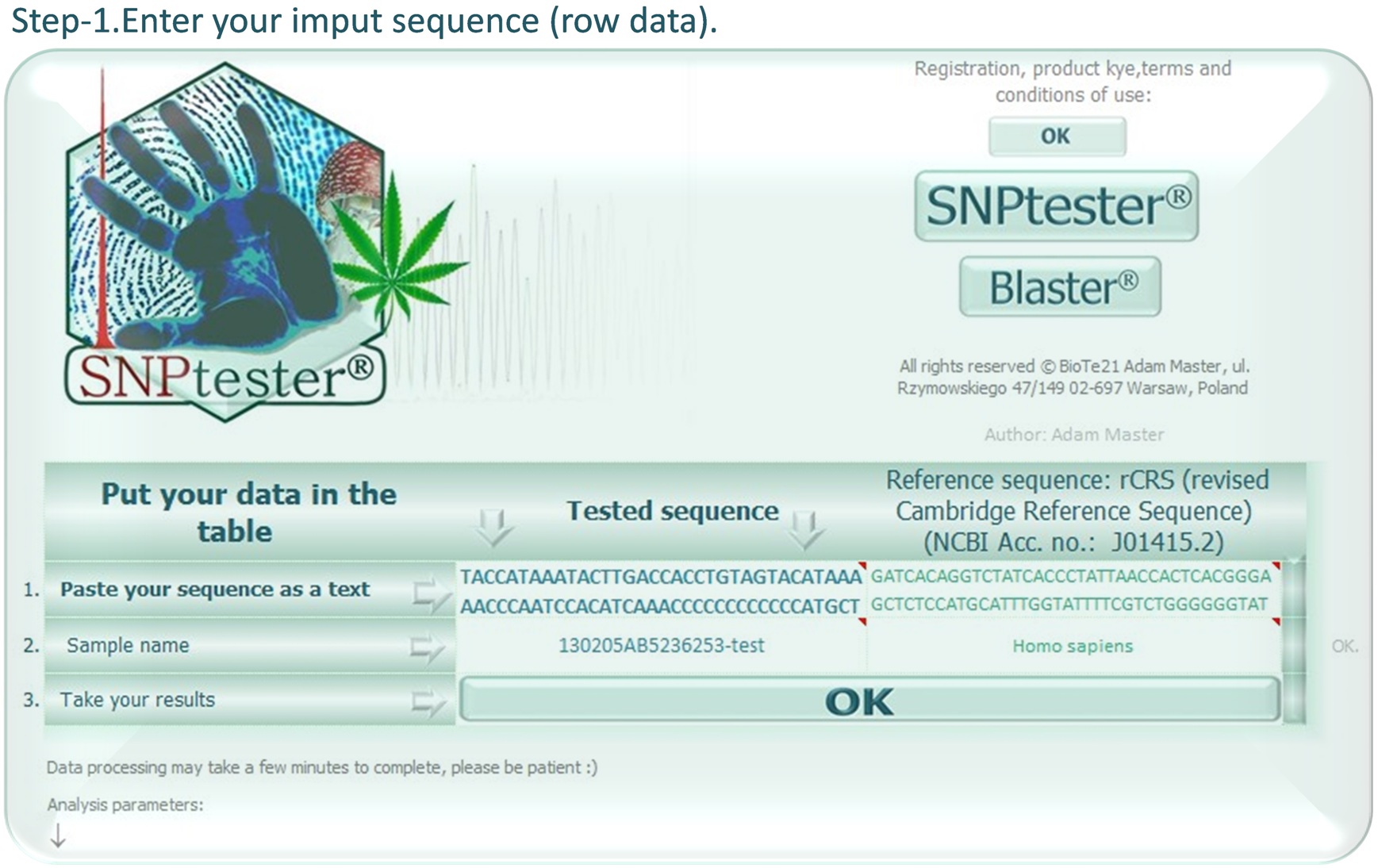
YOU NEED NOT KNOW CORRECT SEQUENCE ANALYSIS AND USAGE OF IUPAC NOMENCLATURE!
The SNPtester® will find and call all sequence variations (SNPs/ mutations) FOR YOU.
Just put your row sequence in the SNPtester® and wait for the results. Click the link below to DOWNLOAD
the current version of the SNPtester® version 2.0
Zip file. Free trials.Test it!
Special free version for scientific and didactic purposes.
Minimum Requirements: Windows XP, Windows 7, Windows 8, RAM: 6 GB Processor 64-bit (x64) prefered Microsoft Excel 2007 or 2010
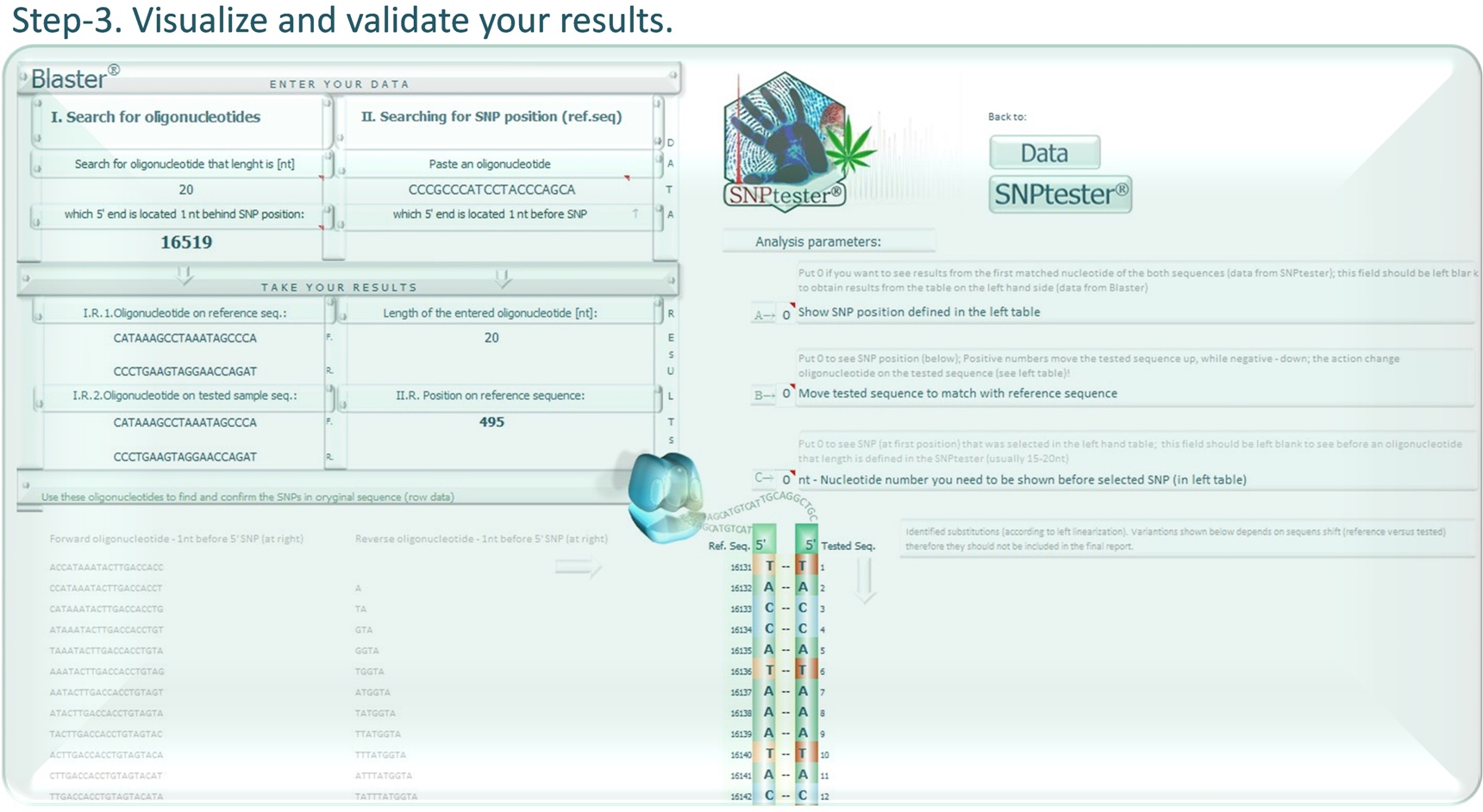
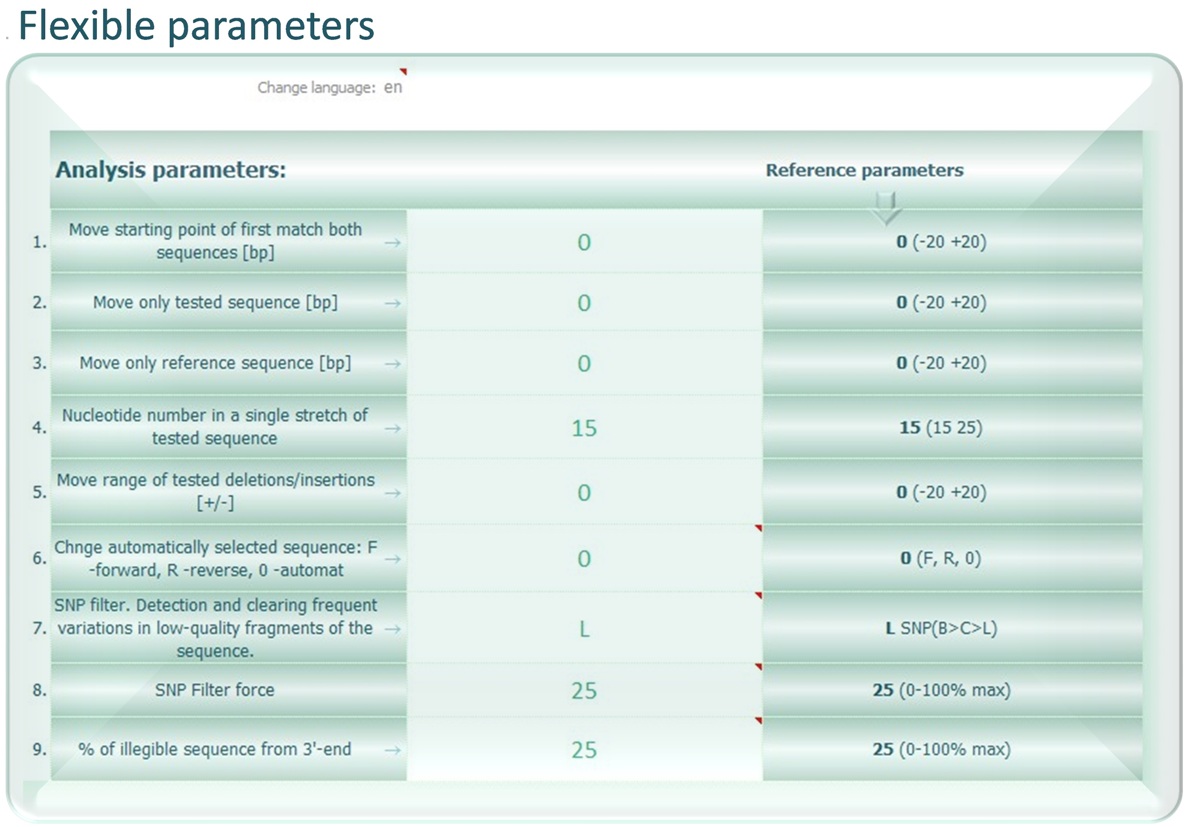
It is possible to enter your reference sequences to the SNPtester® database.
Sequence analysis performed manually was described on mtDNAtest-human.
STR analysis software will be available soon.
 |