This product has been elaborated by Adam Master,
distributed by BioTe21 and presented as follows:
PRINCIPLE
The ITStest Kit offers specially designed master mixes and primers optimized for sensitive and specific amplification of Internal Transcribed Spacer 1 (ITS1) and ITS2 of ribosomal DNA (rDNA) followed by identification of any plant species and subspecies such as Cleome hassleriana phenotypically similar to Cannabis sativa (marihuana), Metasequoia glyptostroboides, Delonis regia, Euterpe oleracea or other fruit trees, seeds and many other plants used in food industry. The Kit allows also for identification of any fungi (foodstuff as well as pharmacological and pathological fungi). Biological materials for the identification are usually of unknown origin, which may be determined with the ITStest-plant & fungi Kit.
The primers of the Kit recognize coding regions of rDNA, that are highly conserved among species but ITS regions are variable due to insertions, deletions, and point mutations (SNPs detected during sequencing). Conserved sequences at coding regions of rDNA allow comparisons of remote species, even between yeast and plants.
The Kit include reagents for PCR systems: all in one standard 0,2uL PCR tube (PCR capsule). You need only an enzyme and common laboratory equipment (thermocycler). If you have an instant access to any sequencer (genetic analyzer), you can perform all laboratory steps in 24h allowing for full identification of your samples in one day.
Advantages of the Kit:
- primers used in the Kit are designed to detect any of plant or fungi species,
- high sensitivity (0.25 pg DNA /reaction),
- specificity and reliability,
- the longest sequence in one amplicon (PCR product) in the biotech market - accuracy,
- excellent quality of results (obtained sequences do not cause problems in the analysis),
- simple and quick implementation of the procedures,
- relatively low cost testing when compared to other methods,
- continuous help during first analyses of results,
- mtDNAtests were validated during international GEDNAP tests. See flowchart below:
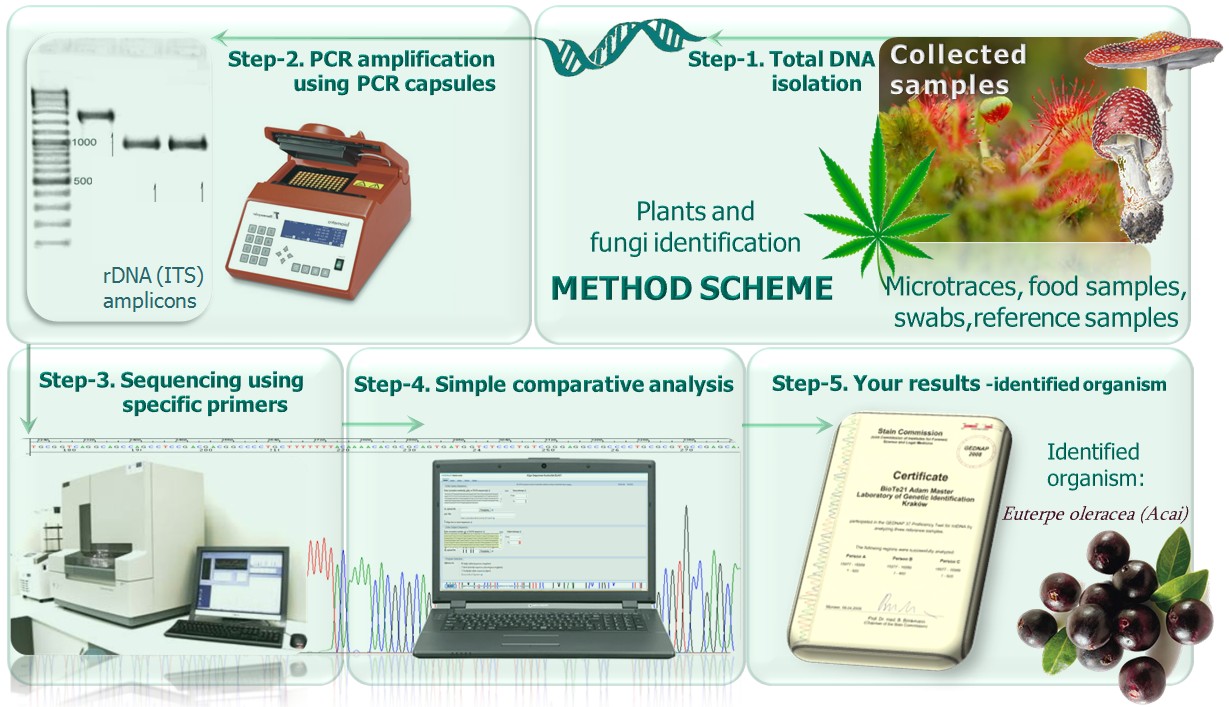
Computational analysis
- Classical method - performed manually, described in mtDNAtest®-human page.
- Using the NCBI BLAST program, the sequence can be compared with the thousands of available sequences previously identified plants. The BLAST provides a list of organisms, which are located in the database. Organisms in the database have a sequence similar to the sequence that we want, obtained data starts with the highest sequence similarity (100% or less), depending on the sequence, and the group of plants we are interested in. A high percentage of similarity suggests a possible identification at the species or subspecies level.
- With our special software: SNPtester® version 2.0, described on seperate page.
Supplied reagents and materials:
Each Kit contains PCR capsules with optimized PCR buffer, standarized PCR primer set, dNTPs and water, as well as control DNA in a separate tube. The capsules require only your sample (DNA) and an enzyme (DNA polymerase).The Kits contains also specific oligonucleotides for sequencing reactions of your PCR amplicons obtained in our PCR capsules.
In addition to the standard primer set allowing for amplification of any plant and fungi species in one capsule, plants-specific primer set and fungi-specific primer set is added to the Kit, that allow selective amplification of fungal or plant sequences.
Equipment, reagents and materials required but not supplied:
PCR enzyme, materials and reagents for agarose gell electrophoresis (validation of PCR products), materials and reagents for sequencing (e.g. BigDyeTerminator Sequencing Kit), thermocycler and access to the genetic analyzer (sequencer). There is also possibility to order that service in our lab. Please contact us to get any answer to questions about the Kits and our services.
Feel free to search through our offer. We will be pleased to answer all your questions concerning examinations – we are here to help! BioTe21 team can help in the analysis and documentation of your results obtained through the use of the ITStest-plant & fungi Kit.
Why ITStest ?
Sequence comparison of the ITS regions is widely used in taxonomy and molecular phylogeny due to:
- the high copy number of rRNA genes easy to amplify even from small quantities of DNA,
- a high degree of variation even between closely related species, that can be explained by the relatively low evolutionary pressure acting on such nonfunctional sequences.
ITS has proven especially useful for elucidating relationships among congeneric species and closely related genera such as in Asteraceae. The internal transcribed spacer has typically been most useful for molecular systematics at the species level, and even within species (e.g., to identify geographic races).
Applications
- Identification of unknown plants or fungi samples (scientific purposes),
- Phylogenetic studies, systematic botany,
- Identification of underground plant parts to species,
- Identification of underground fungi parts to species
(eg. truffle mycelium, mycorrhizal fungi) - Detection of fungal pathogens in plants (agriculture),
- Identification of pathogenic fungi (medical purposes),
- Genetic identification of specimens containing from narcotic crops
- Comparison of fungi DNA profiles obtained from microtraces (e.g. soil samples) and profiles obtained from reference samples (e.g. obtained from a victim's body in a laboratory of forensic medicine),
- Quality control in food processing (food industry),
- Detection of fungal pathogens in food (eg. in crop, food concentrates),
- Safety of foods derived from modern biotechnology.
Scientific Background
rDNA in short
Ribosomal DNA (rDNA) is a DNA sequence that codes for ribosomal RNA (rRNA). Ribosomes are assemblies of proteins and rRNA molecules that translate mRNA to produce proteins. rDNA of eukaryotes consists of a tandem repeat of a unit segment, an operon (polycistronic rRNA precursor transcript), composed of NTS, ETS, 18S, ITS1, 5.8S, ITS2, as well as 28S tracts. rDNA has another gene, coding for 5S rRNA, located in the genome in most eukaryotes.Genes encoding ribosomal RNA and spacers occur in tandem repeats that are thousands of copies long, each separated by regions of nontranscribed DNA termed intergenic spacer (IGS) or nontranscribed spacer (NTS).
The rDNA have low rate of polymorphism (SNP) among species, which allows interspecific comparison to elucidate phylogenetic relationship using only a few specimens. Human 5.8S rRNA has 75% identity with yeast 5.8S rRNA. In cases for sibling species, comparison of the rDNA segment including ITS tracts among species and phylogenetic analysis are made satisfactorily. rDNA can provide phylogenetic information of species belonging to wide systematic levels. ITS (internal transcribed spacer) refers to a piece of non-functional RNA situated between structural ribosomal RNAs (rRNA) on a common precursor transcript. During rRNA maturation, ITS and ETS pieces are excised and as non-functional maturation byproducts rapidly degraded. The ITS region is now the most widely sequenced DNA region in fungi. Because of its higher degree of variation compared to other genic regions of rDNA, variation among individual rDNA repeats can sometimes be observed within both the ITS and IGS regions.
PCR is a method which allows to genotype and identify plants. It is a safe and relatively simple method. Genotyping DNA fragments containing the mutations such as insertions or deletions, usually relay on the direct size analysis of the amplified DNA fragments. In this method the most important are substitutions and the selection of the appropriate primers, allowing for fast and reliable amplification of plant or fungi DNA even when microtraces are investigated.
Small Nucleotide Polymorphisms (SNP)
Differences arising from single nucleotide polymorphism (SNP) genotyping are ideal for plants. They provide a number of markers used by computer programs for the selection of key sets (key crops). Recent improvements to PCR and sequencing techniques of new generation simplify the new generation of genotyping SNPs, which makes the analysis of genotypes is becoming easy and more profitable. SNP are the small changes in the DNA sequence.
The use of plants genotyping in cryminology
The use of plant DNA to determine their genotype is becoming widely used in criminal cases. Genotyping is a method which determines nucleotide sequence of specific DNA regionssuch as gene fragments or whole genes or changes in these regions: polymorphisms, mutations. Distinguishing the difference between genotypes is the most important aim of this molecular technique. Typically, plant identification can be performed using alternative resources, over a precise determination of the separate types of plants is mostly not required. However, the combination of a suspect to a particular plant can be very valuable evidence. There are laboratories which conduct research for clients in need of evidence in the ongoing criminal cases. Research related to criminal matters such as: to distinguish between the type of marijuana without THC and its drug equivalent. The role of molecular systematic botany in forensic medicine is getting wider and more common. Techniques based on DNA analysis are successfully used in criminal cases, and they are used to identify the suspect on the basis of marks secured at the crime scene, or the identification of strains of marijuana. This technology is working on DNA obtained from individual specimens of known and unknown plant species as well as mixtures of plants. A large number of botanical evidence is in the form of the mixture and, as in consequence is not commonly used in the investigations. The possibility of separating and identifying the individual components of mixtures of vegetable can bring additional information about evidence that had previously been ignored or considered too valuable. NCBI Gene bank database contains many plant DNA sequences that can be used to identify the botanical evidence. These sequences include genes, such as rDNA and ITS region. Because of the extensive collection of DNA sequence databases rbcL locus was chosen as a model system for the development of molecular methods for the analysis of trace evidence. One of the rbcL locus complementary in the study of trace evidence includes the ITS region. ITS region of the nuclear genome, located on either side of the region encoding the ribosomal DNA. It is a high copy-number region and has a higher volatility than the rbcL sequences.
Phylogenetic methods may supplement our technology and are based on the model of the evolution of the DNA sequence and are used to establish relationships between the different organisms. These models are being used in programs such as Paup Phylogenetic Analysis Using Parsimony. This method is useful for determining or confirming the genus or family of the specimen to which the sequence. With the equalizing structure of the nucleotide sequence that contains evidence and closely related sequences, a sequence can be classified within the kind of evidence and / or family. The highest amount of information is provided by sequence identification at level of species, however data stored at level of genus or family are also usefull. Sequences recovered from trace evidence can help to identify their geographical origin.
Based on the ecology and plant habitat, which are marked with these sequences, you can request that their geographical origin Tier identify the sample is often dependent on a combination of rarity and recovered plants. This combination of products provides a "description" of the sample, which allows you to adjust or exclude a reference sample to the crime scene and other evidence.
Detection of Cannabis DNA
Drug trafficking is a serious problem, entailing substantial investments made by the customs and excise, and other law enforcement agencies, in order to prevent the importation and supply of illegal substances. Samples of suspect materials must be tested for the presence of illegal substances. Currently, there are several tests that can indicate the presence of Cannabis sativa, including thin-layer chromatography, microscopy and chemical tests topical. More advanced tests include high pressure liquid chromatography or gas chromatography-mass spectrometry.
Here we present the test based on Cannabis sativa genetics, that allows for fast and reliable detection even when microtraces are examinated. The same techniques used by forensic scientists in the human DNA profiling can be used with any biological material containing DNA. Since he was first isolated DNA of Indian hemp (marijuana) it became possible to use the method of DNA profiling to determine the species. Because little is known about the DNA sequence of Cannabis sativa, was not easy to synthesize the primers specific for the amplification of fragments of the Cannabis genome.Our universal as well as Cannabis sativa-specific primers confirmed their usefulness in cannabis examination and detection of small DNA amounts of various Cannabis sativa subspecies.
Literature:
- Hillis DM., Dixon MT. "Ribosomal DNA: Molecular evolution and phylogenetic inference". The Quarterly review of biology 66 (4): 411–453 1991. PMID 1784710.
- Baldwin BG. "Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: An example from the Compositae". Molecular Phylogenetics and Evolution 1 (1): 3–16. doi:10.1016/1055-7903(92)90030-K (1992). PMID 1342921.
- Chen,Y-C, J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, A. P. Limaye, and B. T. Cookson (2001). "Polymorphic Internal Transcribed Spacer Region 1 DNA Sequences Identify Medically Important Yeasts". J. Clin. Microbiol. 39 (11): 4042–4051, 2001. PMID 11682528.
- Peay KG, Kennedy PG, Bruns TD . "Fungal community ecology: a hybrid beast with a molecular master". BioScience 58: 799–810, 2008.G
- Gardes M, and Bruns TD . "ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhiza and rusts". Molecular Ecology. 2 (2): 113–118, 1993. PMID 8180733.
- Jeeva ML, Sharma K, Mishra AK, Misra RS. "Rapid Extraction of Genomic DNA from Sclerotium rolfsii Causing Collar Rot of Amorphophallus". Genes, Genomes and Genomics. 2 (1), 60-62, 2008.
- White, TJ, Bruns T, Lee S., Taylor J. "Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics". In ‘PCR protocols: a guide to methods and applications’. pp. 315–322, 1990.
- Wierzbicki A.T., Jerzmanowski A.: Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation, Genetics. 2005 Feb;169(2):997-1008.
- Epub 2004 Oct 16.
- Linacre A., Thorpe J.: Detection and identification of cannabis by DNA, Forensic science international 1998, 91: 71-76.
- Henry R.J.: Plant genotyping. The DNA finerprinting of plants. Volume I, CABI Publishing 2001.
As other IVD, the Kit is intended for applications utilizing molecular biology methods. This product is neither intended for the diagnosis, prevention, or treatment of a disease, nor has it been validated for such use either alone or in combination with other our products.